Cell metabolism is extremely complex, especially in plants, as they have a specific compartment called the chloroplast. This organelle in the cytoplasm of plant cells conducts photosynthesis, and performs many biosynthetic processes making it essential to the functioning of plant cells.
Extensive research in plant metabolism over the last 50 years provides the foundation for predicting Cell metabolism by modeling. Yet this task requires collating knowledge, currently scattered in various specialized resources (databases, scientific literature, etc.). What's more, this knowledge must be represented in a form that is both human- and computer-readable for mathematical processing. The web-application
ChloroKB, developed over six years by biologists and bioinformaticians at the Cell & Plant Physiology laboratory and at the Large Scale Biology laboratory, both at our institute, now makes this integrated data available to the scientific community.
ChloroKB focuses on the plant
Arabidopsis thaliana—the reference organism in plant research. With
ChloroKB, the scientists can navigate through a network reconstructed manually that includes more than 1,200 proteins, 1,500 metabolites and 800 complexes located in 5 subcellular compartments.
ChloroKB is a large database that provides expert-reviewed data on genes and metabolites, and is also a software that provides relational data to visualize the links between metabolic pathways.
Visual analysis is at the core of
ChloroKB web-application in which biological processes using CellDesigner standards are hierarchized, the different actors in the chloroplast and in the other cellular compartments sorted by shape and color, and the various transports of molecules connected by arrows. The user can interactively explore the interrelationships in the metabolic network, locate the metabolites at the subcellular level and visualize transports or protein assembly processes.
Finally, because of the contextual representation of the metabolic network of Arabidopsis, many elements that are usually absent from databases are represented, such as certain reactions that are still unclear to scientists, or missing steps within the network. ChloroKB leads to the formulation of new biological hypotheses. For example, data assembly reveals new elements such as the co-occurrence of synthesis and degradative processes in the same cell—a co-occurrence that implies the existence of regulatory processes that have not yet been unraveled.
According to researcher Gilles Curien: "
Without ChloroKB, accessing this amount of expert-reviewed data would require weeks of data acquisition even for simple processes" This web application "
facilitates the acquisition of knowledge by neophytes and simplifies communication between experts in the field": ChloroKB thus comes as a very original analytical visual tool and an essential resource for the quantitative and predictive modeling of plant metabolism.
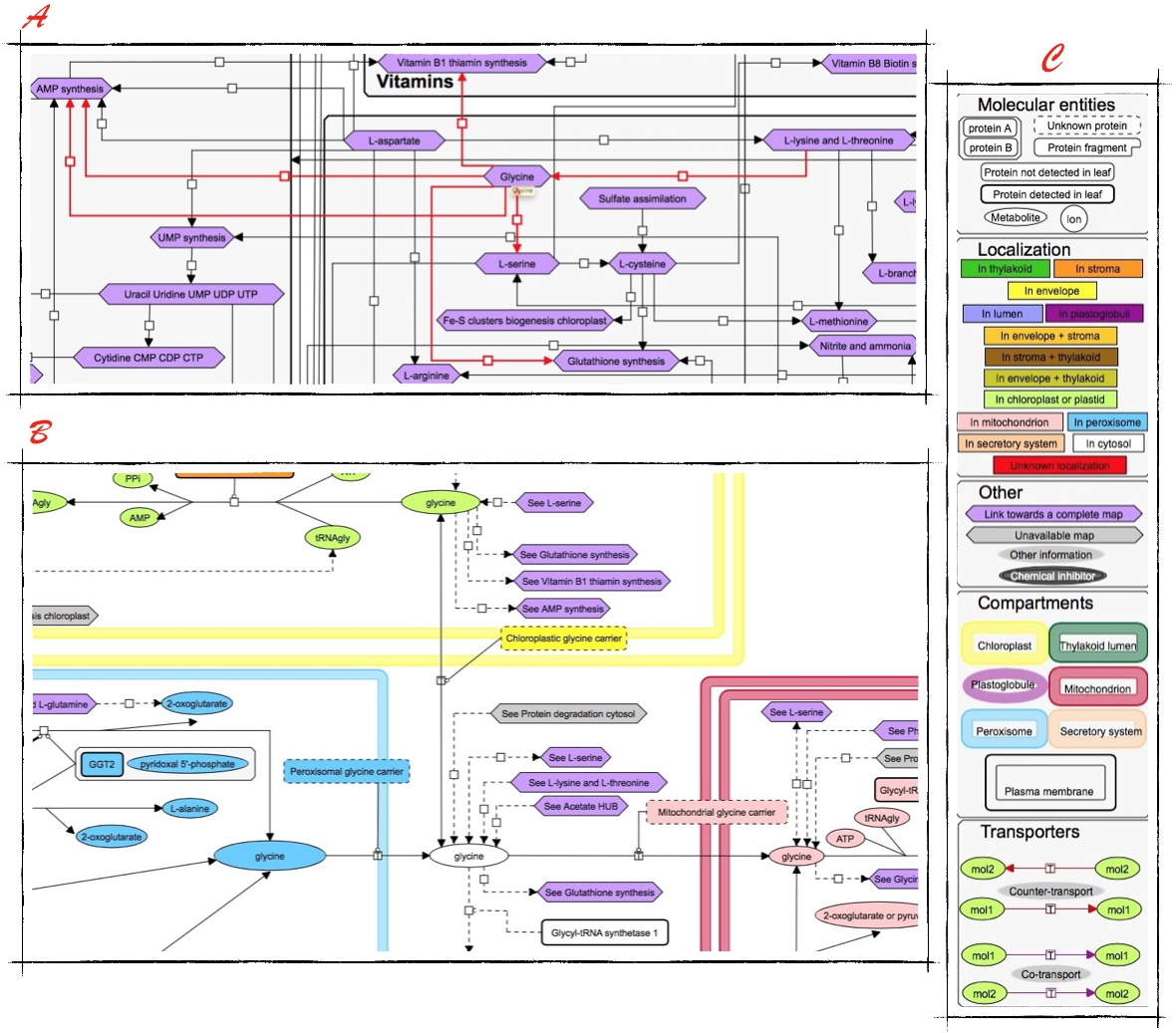
ChloroKB.
A - Preview and zoom of ChloroKB with Glycine taken as an example.
B - Extract of a metabolic map. C - Legend giving an indication of the wealth of ChloroKB.
This work was funded by the GRAL Labex.