 IntroductionPost-translational modifications (PTMs) refer to the covalent processing of a protein after it has been fully translated. PTMs provide enormous chemical and functional diversity (glycosylation, phosphorylation, acetylation, methylation…); they can be reversible or irreversible and are either spontaneous or deposited/removed from targeted residues by specific enzymes (kinases, phosphatases, methyltransferases…). PTMs are known to act alone or in combination to regulate nearly all aspects of protein function (enzyme activity, targeting, turnover rate, interactions with other molecules…). PTMs are involved in the regulation of a wide range of biological processes, including chromatin structure through histones modifications, transcription, translation, metabolism and cell signaling. Studies focusing on PTMs are required to understand how the regulation of protein function is fine-tuned in a constantly changing environment.
IntroductionPost-translational modifications (PTMs) refer to the covalent processing of a protein after it has been fully translated. PTMs provide enormous chemical and functional diversity (glycosylation, phosphorylation, acetylation, methylation…); they can be reversible or irreversible and are either spontaneous or deposited/removed from targeted residues by specific enzymes (kinases, phosphatases, methyltransferases…). PTMs are known to act alone or in combination to regulate nearly all aspects of protein function (enzyme activity, targeting, turnover rate, interactions with other molecules…). PTMs are involved in the regulation of a wide range of biological processes, including chromatin structure through histones modifications, transcription, translation, metabolism and cell signaling. Studies focusing on PTMs are required to understand how the regulation of protein function is fine-tuned in a constantly changing environment. Research activities
Research activities Our research activities aim at characterizing the biotin (vitamin B8) biosynthesis pathway and the covalent attachment of biotin to proteins (biotinylation). Also, we aim at identifying and characterizing non-histone methylated proteins to decipher the role of this PTM in plants.
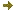 Biosynthesis of biotin and biotinylation of proteins
Biosynthesis of biotin and biotinylation of proteins - We have identified, characterized and localized the four steps involved in the biosynthesis of biotin. We also solved the 3D structure of a bifunctional enzyme of this pathway, which enabled to understand the coordinated action of the two catalytic domains (
Figure 1). Biotin is able to carry carboxyl groups only when covalently attached to biotin-dependent carboxylases. The covalent attachment of biotin to carboxylases is catalyzed by holocarboxylase synthetase (HCS). In plants, we evidenced four different biotin-dependent carboxylase activities localized in the cytosol, chloroplasts and mitochondria. HCS localization in plant cells parallels these activities, suggesting that carboxylases are biotinylated in their compartment of residence. It is likely that a single HCS gene is able to address the protein to the different plant cell compartments and that initiation of the translation is regulated through a micro ORF.
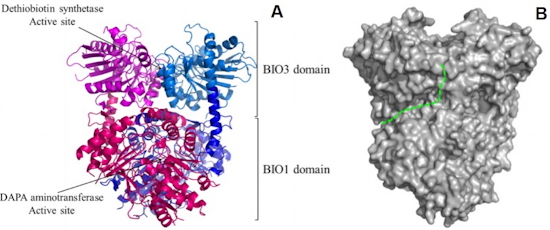 Figure 1: Ribbon and surface representations of the BIO3-BIO1 dimer.
Figure 1: Ribbon and surface representations of the BIO3-BIO1 dimer.
(A) DAPA-aminotransferase (BIO1) and dethiobiotin synthetase (BIO3) domains from each monomer are colored in dark/light blue and red/magenta, respectively.
(B) The external crevice involved in the transfer of the DAPA intermediate is delineated by a dotted green line.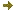 Identification of methylated proteins in chloroplasts
Identification of methylated proteins in chloroplasts - In plants, identification of non-histone methylproteins at a cellular or subcellular scale is still missing. To gain insights into the extent of this modification in chloroplasts we set up a workflow to specifically identify lysine and arginine methylated proteins from proteomic data used to produce the Arabidopsis chloroplast proteome (collaboration with the Large Scale Biology Laboratory (
BGE) in our institute). We could identify more than 20 chloroplastic methylproteins split between the stroma, thylakoids and envelope sub-compartments. They belong to essential metabolic processes, including photosynthesis, and to the chloroplast biogenesis and maintenance machinery (translation, protein import, division) (
Figure 2). We also identified protein methyltransferases targeted to plastids and started to build up the methyltransferases/methylproteins network. These data suggest a role of protein Lys methylation in carbon metabolism in chloroplasts.
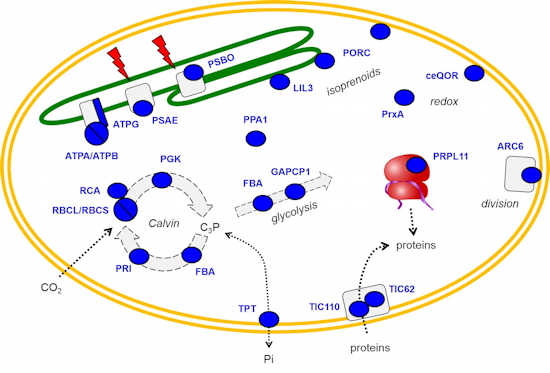 Figure 2: Overview of methylproteins from Arabidopsis chloroplasts.
Figure 2: Overview of methylproteins from Arabidopsis chloroplasts.
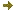 Methylation of ribosomal protein L11 in plastids and mitochondria
Methylation of ribosomal protein L11 in plastids and mitochondria - Methylation of ribosomal proteins has long been described in prokaryotes and eukaryotes but our knowledge about the enzymes responsible for these modifications in plants is scarce. We showed that the protein methyltransferase PrmA from Arabidopsis is dually-targeted to chloroplasts and mitochondria (
Figure 3). Mass spectrometry and enzymatic assays indicated that the enzyme methylates RPL11 in plasto- and mitoribosomes
in vivo. The conservation of PrmA during the evolution of photosynthetic eukaryotes together with the location of methylated residues at the binding site of translation factors to ribosomes suggests that RPL11 methylation in plant organelles could be involved, in combination with other post-translational modifications, in optimizing ribosome function.
 Figure 3: Dual targeting of the methyltransferase PrmA to chloroplasts and mitochondria contribute to the methylation of ribosomal proteins L11 on highly conserved lysine residues.
Figure 3: Dual targeting of the methyltransferase PrmA to chloroplasts and mitochondria contribute to the methylation of ribosomal proteins L11 on highly conserved lysine residues.
 Molecular evolution of the substrate specificity of chloroplastic aldolases/Rubisco lysine methyltransferases in plants
Molecular evolution of the substrate specificity of chloroplastic aldolases/Rubisco lysine methyltransferases in plants - We have previously shown that the methylation of Rubisco and fructose-1,6-bisphosphate aldolases (FBAs), enzymes that are involved in CO
2 fixation in chloroplasts, by the protein methyltransferase LSMT varies in a species-specific manner. FBAs are trimethylated by LSMT in all plant species whereas Rubisco the Rubisco large subunit is trimethylated only in a few species (
e.g. Fabaceae and Solanaceae). By using domain-swapping and site-directed mutagenesis approaches we showed that the His-Ala/Pro-Trp triad located in the central part of LSMTs is the key motif to confer the capacity to trimethylate Rubisco. The distribution of the motif into a phylogenetic tree indicates that the ancestral function of LSMT was FBA trimethylation (
Figure 4). In a recent event during higher plants evolution this function evolved in ancestors of few plant species to include Rubisco as an additional substrate to the archetypal enzyme.
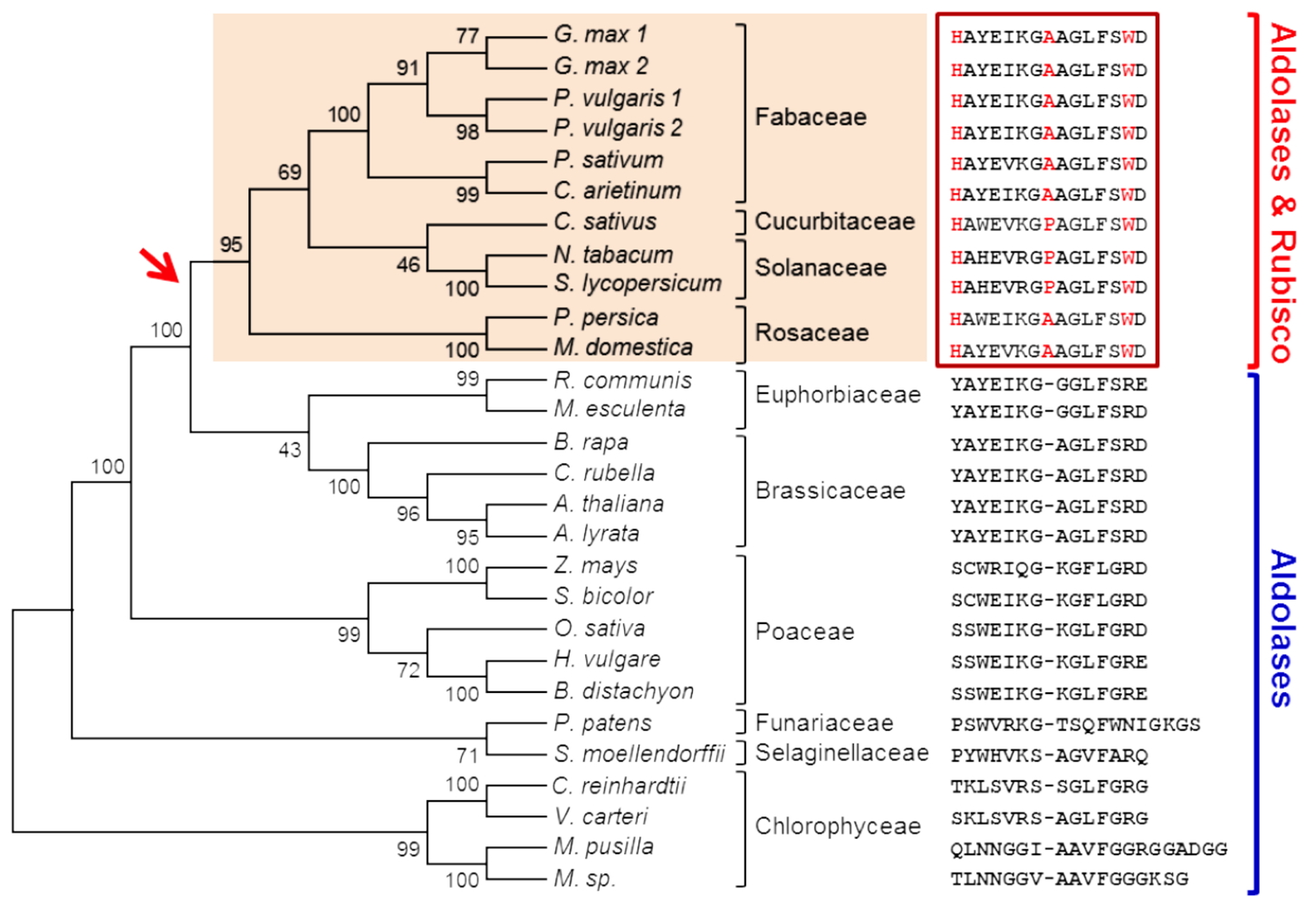
Figure 4: Phylogenetic tree showing the evolution of the substrate specificity of LSMT enzymes. Key words
Key words Biotin; biosynthesis; biotinylation; regulation; methylation; methylated protein; chloroplast; mitochondrion; evolution.