An important contribution to our understanding of cellular mechanisms is the assignment of protein localization in the different subcellular compartments. In eukaryotes, organelles such as mitochondria and chloroplast have a bacterial endosymbiotic origin. The genes of this ancestral bacterium migrated to the cell nucleus and their products, the proteins are synthesized in the cytoplasm. This is why the biogenesis and function of mitochondria or chloroplast involve the migration of these proteins from the cytoplasm into the organelle across the delimiting membrane system. For the most part, these proteins have acquired a signal sequence called import sequence that allow them to be properly addressed in the sub-cellular compartment. To predict the presence of an import sequence (also called transit peptide in the literature) is a key component for the annotation of subcellular localization of a protein. However, existing tools were developed using animals or terrestrial higher plants sequences, and thus prove to be unsuitable for the Green Algae. In this new study, the goal of the Exploring the Dynamics of Proteomes (EDyP) at the Large Scale Biology Laboratory was to design a software dedicated to green algae, called PredAlgo
[1].
The unicellular green algae
Chlamydomonas reinhardtii was chosen as a source of experimental data for training the software. It is indeed the only green algae for which there is a large enough inventory of proteins for which subcellular localization is known. Such proteins have their import sequence cleaved during their transfer into the chloroplast or mitochondria. The precise identification of import sequences (
i.e. at the cleavage site) of these proteins could be obtained by re-analyzing data from tandem mass spectrometry (
Figure 1) previously acquired for the analysis of the mitochondrial
[2] and chloroplastic proteomes
[3].
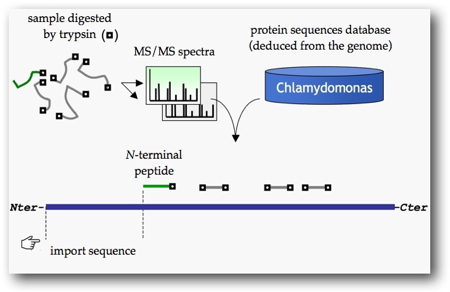
The analysis of sequences adjacent to the identified cleavage sites reveals general trends (
Figure 2) which does not allow to obtain clear and distinctive consensus for the chloroplastic or mitochondrial compartments. As a result, PredAlgo was developed on the principle of "neural networks", a black box in which training optimizes decision rules that are not interpretable by humans.
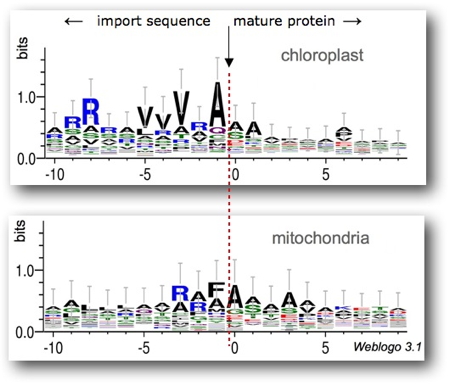
Figure 2: Relative occurrence of amino acids over a window of 10 positions apart from the cleavage site (arrow): these schemes have been generated on the basis of the chloroplastic and mitochondrial proteins used for the training of PredAlgo.
In comparison with existing software, PredAlgo achieves the best performance for algae, especially for the discrimination between chloroplastic and mitochondrial proteins. The tool is relevant not only in Chlamydomonas but also for other close lineages of green algae. It is less predictive in the case of the localization of mitochondrial proteins from most distant algae.
Green algae are increasingly recognized as a potential source of 3rd generation biofuels (fatty acids, sugars, hydrogen) whose production is associated with the chloroplast metabolism. PredAlgo is the best suitable tool for predicting subcellular localization in these organisms and should accelerate the understanding of their metabolism and compartmentalization in order to increase the knowledge base required for green algae engineering.