Research themes of the team
Published on 17 November 2025
Body text 1
 Light signaling, utilization and dissipation
Light signaling, utilization and dissipation
In photosynthetic organisms light is a signal perceived by photoreceptors,
i.e. proteins that convert light radiation into biological signals regulating essential cell functions (i.e. stomata opening and closure, photo-orientation of chloroplasts, plant development). Light is also the energy source used to convert CO2 into organic metabolites. When light is absorbed in excess to the photosynthetic capacity of the cells, photoprotective mechanisms known as NPQ (non-photochemical quenching,
Figure 1) are activated in order to reduce photodamage.

Figure 1:
Chlorophyll (Chl) molecules after absorbing light are excited to the singlet excited state (Chl*). They can relax back to their fundamental state (Chl) either by photochemistry (photosynthesis), or by light reemission (fluorescence) or by non radiative dissipation (heat). The latter process allows dissipating excess absorbed light, thereby reducing production of harmful species like ROS. Light absorption is also regulated by State Transitions, a process involving a « slow » movement of light absorbing proteins between the two photocenters of oxygenic photosynthesis in order to optimize their use of absorbed light. In general, state transitions mostly occur at limelight intensities limiting photosynthetic activity, while NPQ is mostly active at oversaturating photon fluxes.
NPQ can be divided in mainly two components:
(1) qE (high-energy state quenching), the major and fastest component of NPQ that depends on the buildup of a high ΔpH across the thylakoid membrane (i.e., a low lumen pH), on specific carotenoids and, in the case of Chlamydomonas, on the protein LHCSR3. It is a nucleus-encoded, chloroplast-localized protein that is induced under strong illumination. Full induction of LHCSR3 and qE takes few hours at variance with higher plants that the protein effectors of qE are constitutively expressed (Allorent
et al.,
2013). The signal transduction process that controls the expression of LHCSR3 involves Ca2+ signaling events and requires an active photosynthetic electron flow (Petroutsos
et al.,
2011) but the molecular mechanism behind this process in still largely unknown.
(2) state transitions (qT), a change in the relative absorption capacity of the two photosystems (PSII and PSI). (Figure 2). This results from reversible phosphorylation of light harvesting proteins. We have recently shown that the increased ability of Chlamydomonas to perform state transitions compensates for the slow induction of qE during the early stages of high light acclimation (Allorent
et al.,
2013, Nagy
et al.,
2014).
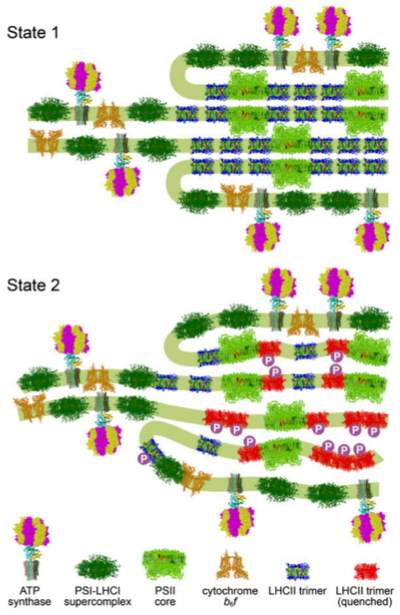
Figure 2:
Chloroplast remodeling during state transitions in
Chlamydomonas reinhardtii as revealed by noninvasive techniques in vivo.
Hypothetical model for the chloroplast remodeling during state transition in C. reinhardtii. Side views of the membrane planes showing alterations in the thylakoid ultrastructure and photosystem supercomplex composition. For S1 (Upper), thylakoids are more stacked, and large arrays of PSII–LHCII supercomplexes reside in the appressed regions. For S2 (Lower), a number of LHCII proteins are phosphorylated, and the thylakoids are partially unstacked and undulated.
Project:
We have recently discovered that a photoreceptor protein controls transcription of LHCSR3 and that LHCSR3 transcription is inhibited by acetate and other metabolites. We are currently trying to unveil the mechanisms that tightly regulate the interconnection between photo-perception, photo-utilization (metabolism) and photo-protection in the green microalga Chlamydomonas. Part of
this research is funded by the French National Research Agency (ANR-10-LABX-49-01; Labex GRAL).
 Redox regulation of electron flow-Ions and photosynthesis
Redox regulation of electron flow-Ions and photosynthesis
Most biochemical reactions in living cells are fuelled by adenosine triphosphate (ATP) and/or the reducing agent reduced nicotinamide adenine dinucleotide phosphate (NADPH). In plant cells, these components are generated in mitochondria and chloroplasts through the respiratory and photosynthetic electron transfer chains. Nearly 50 years ago, Mitchell proposed a unifying mechanism through which the respiratory and photosynthetic electron transfer reactions are coupled to proton pumping across the inner mitochondrial and thylakoid membranes; the thylakoid membranes are located within chloroplasts and contain the protein-pigment complexes that catalyze the primary reactions of photosynthesis (Figure 3).
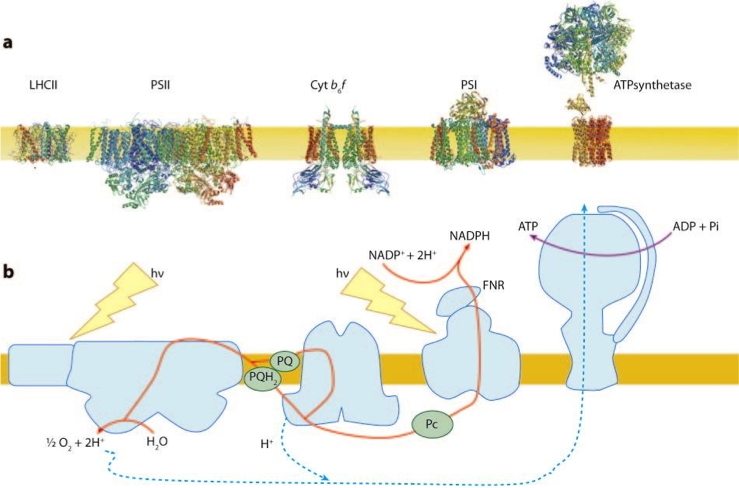
Figure 3:
a - Transmembrane organisation of the major photosynthetic proteins in their native oligomerization state.
b - Schematic representation of the pathway for photosynthetic electron flow.
This proton movement generates a transmembrane electrochemical gradient called the proton motive force (pmf), which is comprised by a proton gradient (ΔpH) and an electric field (ΔΨ). While both components of the pmf are equally necessary for ATP synthesis, the ΔpH specifically regulates photosynthetic control and non-photochemical quenching (NPQ,
Figure 1) by modulating both the protonation state of key regulatory proteins and the rate of electron flow in the cytochrome b6f complex. It is therefore assumed that the size of the ΔpH must be regulated in response to environmental stimuli through a still elusive mechanism. Our previous study suggests that potassium flux across the thylakoid membrane through an ion channel (TPK3 Checchetto
et al.,
2012, Carraretto
et al.,
2013) regulates the proton motive force by counterbalancing the H+ gradient with K+ flux and thus impacts photoprotection by modulating non photochemical quenching (NPQ) and electron flow, ultimately affecting plant fitness.
Questions:
Is it possible to optimize photosynthesis fluxes in algae (Chlamydomonas reinhardtii), mosses (Physcomitrella patens,
Marchantia polymorpha) and plants (Arabidopsis thaliana) by regulating ion fluxes? To answer this question, we are using a comprehensive and integrative approach, to identify new players and dissect the coordinate regulation of light conversion into chemical energy. To this aim we are combining our expertise with competences of our collaborators (electrophysiology, bioinorganic chemistry and molecular genetics) to identify putative chloroplast ion channels, generate down-regulated mutants, characterize their function
in vitro (electrophysiology) and
in vivo (using a home-made biophysical tool: Stark spectroscopy) and collate their phenotype to ion dynamics
in vivo (by development and application of new fluorescent sensors for imaging metal ion fluxes in living plant cells and whole organisms). This
research is funded by a Human Frontier Science Program (HFSP), which started in September 2015 and gathers a consortium of laboratories from Japan (University of Kyoto), USA (University of Berkeley) and Europe (University of Padova, Italy and our team).
 Role of organelle interaction in optimizing carbon assimilation
Role of organelle interaction in optimizing carbon assimilation
In the course of evolution photosynthetic organisms developed sophisticated molecular regulatory mechanisms allowing optimization of light capture and distribution of the energy temporally stored in carbon/redox compounds. The molecular details and the integrated functioning of these dynamical systems is still not well understood. Improvement on our understanding requires to fill in the bridge between abundant (but still insufficient) molecular details, communication between mitochondria (the motor) and chloroplast (the solar panel) and systems behavior at the cellular level. Our recent data (Bailleul
et al.,
2015) indicate that this mechanism is particularly relevant in diatoms,
i.e. organisms that have been dominating photosynthetic marine eukaryotes in the last 35 millions of years. In diatoms, regulation of the ATP/NADPH ratio for proper carbon assimilation occurs through extensive energetic exchanges between plastids and mitochondria (Figure 4). This interaction comprises the rerouting of reducing power generated in the plastid towards mitochondria and the import of mitochondrial ATP into the plastid, and is mandatory for optimized carbon fixation and growth. We propose that the process may have contributed to the ecological success of diatoms in the ocean (see our highlights).

Figure 4:
Representation of a diatom and of the cellular mechanisms coupling photosynthesis and respiration in that organism. The chloroplast, producing ATP and NADPH thanks to light, is closely associated to the mitochondria where respiration occurs. These two sub-cellular compartments can thus exchange the ATP or the NADPH molecule that they produce optimizing CO2 fixation in diatoms.
Questions:
In this project we want to investigate the mechanisms regulating the energy fluxes. Unlike a power station, which supplies only one form of energy, electricity, the chloroplast must supply both ATP and NADPH in precisely the right proportion to match consumption. The ATP/NADPH ratio and its control set into place by photosynthetic organisms during evolution under natural evolutionary pressure is expected not to be optimal for biotechnological applications. In order to manipulate the ATP/NADPH ratio we have started identifying the molecular actors responsible for the synthesis (e.g. NAD+ kinase and its control), and exchange (i.e. the malate shuttle in diatoms) of reducing power in plants and algae. Our goal is to build a comprehensive model of energy fluxes in the plant cell starting from a detailed description of metabolism in algae. For this we will extend our expertise in modelling metabolism from the model plant
Arabidopsis thaliana (Curien
et al.,
2009, Curien
et al.,
2014) to other organisms using metabolic reconstructions (Figure 5) and modelling in collaboration with
EDyP team (our institute, BGE laboratory, CEA-Grenoble) and partners from abroad (O. Ebenhöh, Ebenhöh
et al.,
2014,
Figure 6), in the frame of a Marie Curie ITN project (AccliPhot project), which includes 12 partners from six different European countries.
Figure 5:
Reconstruction of metabolic networks. The understanding of multi-compartimented processes involving thousands of compounds (proteins, metabolites, membranes) and multi-level controls (phosphorylation, allosteric controls, transport processes) requires the development of new visualisation tools. Such bioinformatics tools are useful to generate new hypotheses and interpret data but also to communicate and transmit knowledge across generations. The experience acquired on the reconstruction of
A. thaliana metabolism (see
ChloroKB website) will be re-invested for in silico reconstruction of algae metabolism dedicated to a detailed representation of energetic processes.
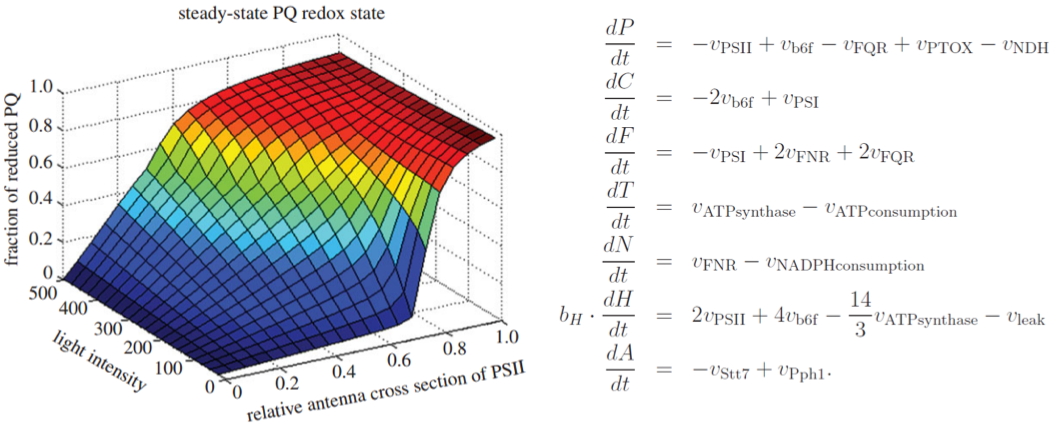
Figure 6:
Short-term acclimation of the photosynthetic electron transfer chain to changing light: A mathematical model Ebenhöh
et al., 2014.
We are also developing biotechnology application of our studies to improve algal biomass productivity, in collaboration with the French SME Fermentalg.
Top page