Uranium (U) is a naturally occurring radionuclide that can accumulate locally in soil and water where it poses potential risks to ecosystems, agrosystems and ultimately human health. Indeed, U is a non-essential element that can be absorbed from the environment and induce both chemotoxic and radiological effects. Although the toxic effects of U are increasingly well characterized, the pathways of uptake of this radionuclide have never been clearly described in either prokaryotic or eukaryotic organisms.
Researchers at our laboratory have identified several mechanisms that allow U to enter the cells of two model eukaryotic organisms, the yeast
Saccharomyces cerevisiae and the plant
Arabidopsis thaliana. In a first step
[1] they showed that U transport in yeast was dependent on the metabolic activity of the cell: U does not enter the cell passively. To identify potential pathways for U uptake, they performed competition experiments with essential metals for yeast. These experiments consist of measuring the effect of increasing concentrations of an essential metal in the extracellular medium on the ability of cells to incorporate U (knowing that a competing metal will reduce U incorporation in a dose-dependent manner). Thus, they identified calcium, iron and copper pathways as potential pathways for U uptake. To provide definitive proof, they analyzed several mutants affected in metal transport and revealed the involvement of the
calcium channel named Mid1/Cch1, as well as of a transmembrane protein, the iron(III) permease Ftr1, in U uptake. Finally, expression of the
Mid1 gene in the ΔMid1 deletion mutant (yeast in which the
Mid1 gene essential for calcium channel formation was deleted) restored the U uptake levels of the wild-type strain, highlighting the central role of this calcium channel in the U uptake process.
Uranium uptake pathways in yeast.
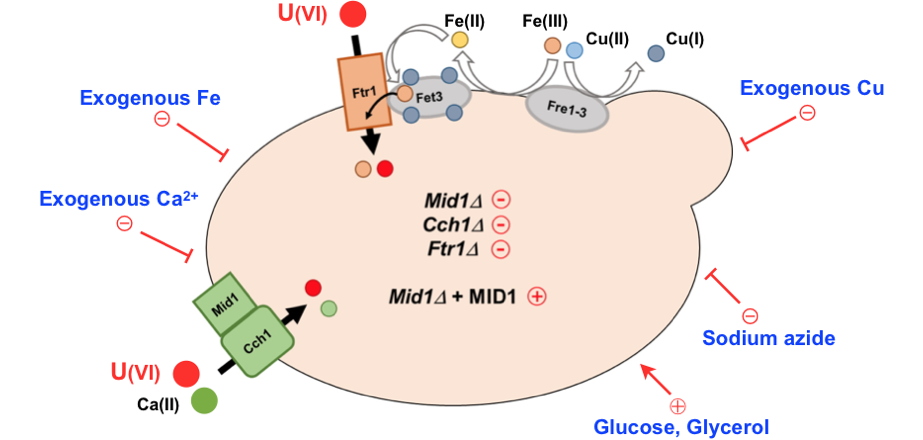
Uranium uptake into the yeast Saccharomyces cerevisiae requires an energy source (glucose or glycerol), a functional respiratory chain (blocked by sodium azide) and involves the high-affinity calcium channel Mid1/Cch1 as well as the ferric iron permease Ftr1.
What about plants? A second study
[2] conducted in parallel by this team provided several pieces of evidence that U is taken up by the roots of the plant
Arabidopsis thaliana via calcium channels. A nutritional deficiency in calcium, is able to induce an increase (1.5 times) in the capacity of the plant to accumulate U in its roots. To confirm the link between U and calcium homeostasis, the researchers showed that U accumulation in roots was inhibited by exogenous calcium (competition phenomenon) and by ionic or chemical calcium channel blockers. These experiments also suggest that different types of Ca
2+-permeable channels, which are very numerous in plants, serve as a pathway for U uptake. Finally, U accumulation in the roots of two calcium channel-deficient
Arabidopsis mutants was found to be strongly reduced, confirming that they directly contribute to U uptake.
Uranium uptake pathways in Arabidopsis roots.
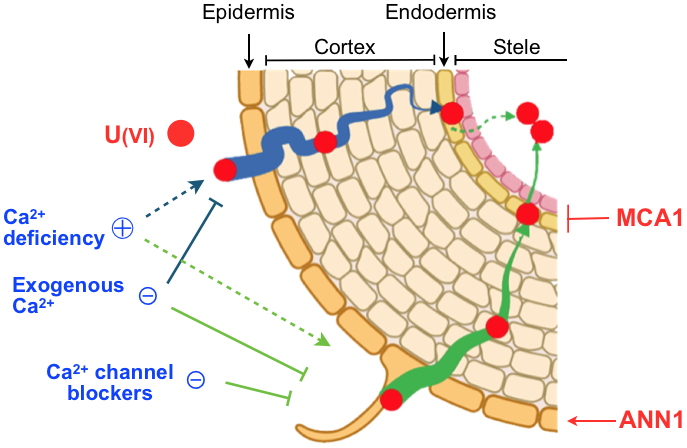
In the roots of Arabidopsis thaliana, U can be transferred to the endodermis through the cell walls (apoplastic pathway, in blue) before being blocked by structures known as Casparian strips. The transfer of U into endodermal cells (symplastic pathway, in green) involves calcium channels, of which the MCA1 protein. The symplastic pathway allows direct radial transfer of U from the soil, with the atypical calcium channel ANN1 participating in the uptake into epidermal cells.
Taken together, these results describe for the first time the U entry pathways in two model eukaryotic organisms. They demonstrate the involvement of calcium channels and iron(III) permease in the cellular uptake of U. This work also paves the way for a simple and rapid identification of potential U transporters from complex multicellular organisms, by functional expression in yeast (unicellular organism), prior to their validation and detailed study in their native organisms.
This work was funded by the nuclear toxicology program of the CEA and by the ANR (GreenU project).
A
calcium channel is an ion channel (cationic to be precise), formed of proteins and crossing the plasma membrane of cells. It allows the passage of calcium ion from the outside to the inside of the cell. The calcium channels involved are
MCA1 and
annexin 1 (ANN1). MCA1 (Mid1-Complementing Activity) is a calcium channel located in the plasma membrane of plant cells. Annexins are proteins that bind to membranes in a calcium-dependent manner and act as atypical calcium channels.