Inside of eukaryotic cell nuclei, chromosomes are more or less compacted, along so-called "chromatin states" that condition the expression of genes they embed. Modifications of these chromatin states, targeted on thousands of genes, are a draining force for development in eukaryotes. Indeed, the differentiation of cells in various types is finely controlled by chemical modifications or "epigenetic marks", brought to the chromatin in a precise and very dynamic manner. These marks regulate access to genes for the transcription machinery, with permissive marks giving access and repressive marks preventing access. The mechanisms that regulate the dynamics of these marks and thereby build the individual body pattern remain poorly understood..
Researchers of the Floral Regulators team in the Cell & Plant Physiology Laboratory [Collaboration] have identified the function of three Jumonji-type enzymes that ensure selective and timely release of repressive marks1 at thousands of genes. The researchers show that the demethylase "RELATIVE OF EARLY FLOWERING 6" and its two close homologues, determine the spatial limits of these repressive marks
1 in the plant genome. Targeted mutagenesis of the three enzymes reveals their redundant and pleiotropic
2 role in organogenesis, impacting vegetative development, growth, flowering and reproduction. Thus, these Jumonji enzymes delimit the repressive domains of chromatin but also contribute to the specific de-repression of genes by direct and indirect targeting mechanisms, via other proteins capable of binding the DNA in a specific manner, depending on the tissue and developmental stage of the plant.
This study sheds new lights on the functions that finely regulate chromatin structure via removal and restriction of epigenetic marks, and opens new perspectives in the understanding of the mechanisms that activate developmental genes.
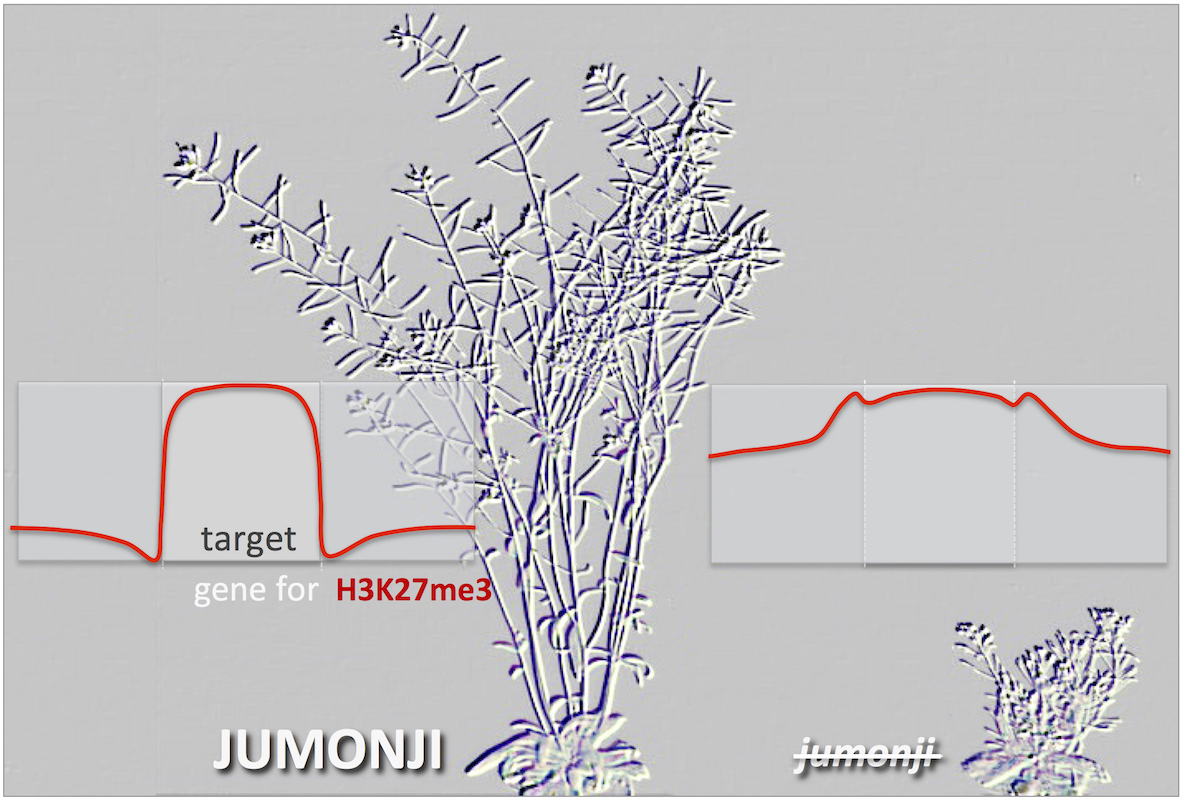
Mean distribution of the H3K27me3 chromatin repressive mark on its target genes and adjacent regions in plants containing the functional JUMONJI enzymes (left) or lacking the 3 specific H3K27me3 demethylases (jumonji, right). The corresponding plants are represented in the background as engraving.
Collaboration: Department for Plant Cell and Molecular Biology, Institute for Biology (Humboldt-Universität Berlin, Germany); Cell & Plant Physiology Laboratory (Université Grenoble Alpes, France); National Key Laboratory of Crop Genetic Improvement (Huazhong Agricultural University, China); Max Planck Institute of Molecular Plant Physiology (Potsdam, Germany); Institute Plant Science Paris-Saclay (Université Paris-sud and Université Paris-Saclay, Orsay, France).
1 These
repressive marks correspond to posttranslational modifications of trimethyl type, carried by Lysine 27 of Histone 3 (H3K27me3)
2 Pleiotropic: which affects several functions.